Interactive jobs
From HP-SEE Wiki
Kozlovszky (Talk | contribs) (→Deep sequencing for short fragment alignment (DeepAligner)) |
|||
| Line 30: | Line 30: | ||
= Deep sequencing for short fragment alignment (DeepAligner) = | = Deep sequencing for short fragment alignment (DeepAligner) = | ||
| - | ''Section contributed by SZTAKI (DeepAligner application)'' | + | ''Section contributed by SZTAKI & OU(DeepAligner application)'' |
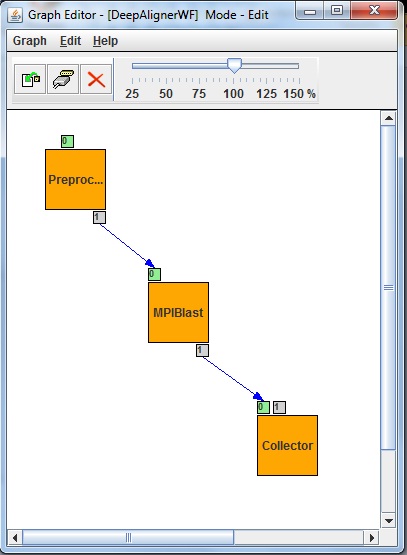
| - | The DeepAligner application’s workflow has been developed as a Parameter Study workflow with usage of autogenerator port (second small box around left top box in Fig xs1.) and collector job (right bottom box in Fig. xs1). The preprocessor job generates a set of input files from some pre-adjusted parameter. Then the second job (middle box in Fig. xs1) will be executed as many times as the input files specify. The second job is an MPI based BLAST executable (MPIBlast) which aligns short sequences. The inputs of the MPI job are the sets of sequences (defined by the researcher) and the already deployed sequence database fragments. The last job of the workflow is a Collector which is used to collect several files and then process them as a single input. Collectors force delayed job execution until the last file of the input file set to be collected has arrived to the Collector job. The workflow engine computes the expected number of input files at run time. When all the expected inputs arrived to the Collector it starts to process all the incoming inputs files as a single input set. Finally output files will be generated, and will be stored on a Storage Element of the DCI shown as little box around the Collector in Fig | + | The DeepAligner application’s workflow has been developed as a Parameter Study workflow with usage of autogenerator port (second small box around left top box in Fig xs1.) and collector job (right bottom box in Fig. xs1). The preprocessor job generates a set of input files from some pre-adjusted parameter. Then the second job (middle box in Fig. xs1) will be executed as many times as the input files specify. The second job is an MPI based BLAST executable (MPIBlast) which aligns short sequences. The inputs of the MPI job are the sets of sequences (defined by the researcher) and the already deployed sequence database fragments. The last job of the workflow is a Collector which is used to collect several files and then process them as a single input. Collectors force delayed job execution until the last file of the input file set to be collected has arrived to the Collector job. The workflow engine computes the expected number of input files at run time. When all the expected inputs arrived to the Collector it starts to process all the incoming inputs files as a single input set. Finally output files will be generated, and will be stored on a Storage Element of the DCI shown as little box around the Collector in Fig 1. |
[[File:Blast_wf.jpg]] | [[File:Blast_wf.jpg]] | ||
| - | Fig | + | Fig 1: DeepAligner workflow with the MPI based Blast job in the middle |
| - | + | More information: [http://wiki.hp-see.eu/index.php/DeepAligner] | |
= In-silico Disease Gene Mapper = | = In-silico Disease Gene Mapper = | ||
Revision as of 23:21, 24 April 2012
CMSLTM @IMBB
Jobs were submitted to the PBS queue manager
Example script:
#!/bin/bash #PBS -q lifesci #PBS -l nodes=2:ppn=5 #PBS -l walltime=10:00:00 # PBS_O_WORKDIR=/home/gkastel/layer_V1/experiment/ # PBS_NODEFILE="/home/gkastel/layer_V1/experiment/nodelist.txt" MPIEXEC="/usr/mpi/gcc/openmpi-1.4.3/bin/mpiexec" cd $PBS_O_WORKDIR NPROCS=10 NRNIV=/home/gkastel/src/nrn-7.1/x86_64/bin/nrniv NLIBS=/home/gkastel/layer_V1/mechanism/x86_64/.libs/libnrnmech.so date +%s > times.txt echo >> times.txt $MPIEXEC -np NPROCS $NRNIV -dll "$NLIBS" -mpi finalpar.hoc date +%s >> times.txt
Invocation:
qsub script-name.sh
Deep sequencing for short fragment alignment (DeepAligner)
Section contributed by SZTAKI & OU(DeepAligner application)
The DeepAligner application’s workflow has been developed as a Parameter Study workflow with usage of autogenerator port (second small box around left top box in Fig xs1.) and collector job (right bottom box in Fig. xs1). The preprocessor job generates a set of input files from some pre-adjusted parameter. Then the second job (middle box in Fig. xs1) will be executed as many times as the input files specify. The second job is an MPI based BLAST executable (MPIBlast) which aligns short sequences. The inputs of the MPI job are the sets of sequences (defined by the researcher) and the already deployed sequence database fragments. The last job of the workflow is a Collector which is used to collect several files and then process them as a single input. Collectors force delayed job execution until the last file of the input file set to be collected has arrived to the Collector job. The workflow engine computes the expected number of input files at run time. When all the expected inputs arrived to the Collector it starts to process all the incoming inputs files as a single input set. Finally output files will be generated, and will be stored on a Storage Element of the DCI shown as little box around the Collector in Fig 1.
Fig 1: DeepAligner workflow with the MPI based Blast job in the middle More information: [1]
In-silico Disease Gene Mapper
Section contributed by SZTAKI
The in-silico Disease Gene Mapper (DiseaseGeneMapper) was ported successfully with the workflow based gUSE development environment. The ported DiseaseGeneMapper application is operated as an on-line service on the HP-SEE’s Bioinformatics eScience Gateway. The life-cycle of the developed workflow was the following:
Figure 1. Porting steps of the application
The graph (or structure) of the workflow was created. This so-called abstract workflow can be used to generate various concrete workflows. The concrete workflows generated from a certain graph can be different concerning the executable code associated with the nodes of the workflow, input and output files associated with ports, specification of the DCI, etc. Using the abstract workflow, a concrete workflow can be generated on-the-fly by configuring detailed properties (first of all the executable, the input/output files and the target DCI) of the nodes representing the atomic execution units of the workflow. After all the properties of the workflow have been set, the workflow is ready to be submitted resulting in an instance of the workflow. A concrete workflow can be submitted arbitrary amount of times and every submission will result in a new instance of the same concrete workflow. For the front-end GUI of the DiseaseGeneMapper application the Application Specific Module (ASM) has been used. The interactive portlet is able to collect application specific information from the researcher (shown Fig xc1).
GUI of the In-silico Disease Gene Mapper application