Libraries usage
From HP-SEE Wiki
Contents |
mpiBLAST
Section contributed by SZTAKI & OU
mpiBLAST is a sequence alignment application in bioinformatics. the software is freely available, open-source, and basically it is a parallel implementation of NCBI BLAST. By efficiently utilizing distributed computational resources through database fragmentation, query segmentation, intelligent scheduling, and parallel I/O, mpiBLAST improves NCBI BLAST performance by several orders of magnitude while scaling to hundreds of processors. mpiBLAST is also portable across many different platforms and operating systems. In the applications developed by SZTAKI and OU we have used mpiBLAST-1.6.0 compiled from source on each of the supercomputers.
BLAST
BLAST is tool which searches a large database of known nucleotide or peptid sequences for similar strings to the query sequence provided by the user. The query sequence can either be a nucleotide or a peptid sequence. The BLAST tool supports both types of queries, and it can even translate one type of query to the other on the fly. The program supports multiple search types: blastn, blastp, blastx, tblastn, tblastx
mpiBLAST requirements mpiBLAST requires the NCBI toolbox (provided with the mpiBLAST sourcecode) and an MPI implementation. For the HPSEE project we are using Open-MPI.
Installation
mpiBLAST was installed on all available NIIF supercomputers following the Installation Guide on the project's website (http://www.mpiBLAST.org/Docs/Install). One minor modification was required because the ncbi toolkit did not compile due to an unresolved dependency issue. After some debugging the reason for this turned out to be the motif package which was installed on the machine, but it could not be used for some reason. As this package would only be required by mpiBLAST if a graphical user interface was present (which is not the case for the supercomputers), motif support was disabled manually for the ncbi toolkit (<mpiBLAST-source>/ncbi/make/makedis.csh - SET HAVE_MOTIF=0). For detailed instructions on installing mpiBLAST please refer to the installation guide.
mpiBLAST in the HPSEE applications developed by SZTAKI mpiBLAST is the core application of both portlets developed in the HPSEE project by SZTAKI, DiseaseGeneMapper and DeepAligner. We use it find short protein or nucleotide sequences in NCBI databases.
Preparing the databases The NCBI databases are stored locally on the shared storage of the supercomputers. Both programs check if the database specified can be found on the servers, and downloads them automatically if they are missing.
The databases can be downloaded from an ftp site either using wget or a simiar tool, or it is possible to use to database updater tool NCBI provides which handles missing files, updated remote db and such things automatically. The tool can be found at: http://www.ncbi.nlm.nih.gov/blast/docs/update_blastdb.pl In our applications we use the update_blastdb.pl tool.
On the NCBI website the databases are stored in their original format which has to be converted first to the mpiBLAST database format using the command mpiformatdb. The syntax of mpiformatdb is the following:
[login.budapest:~/mpiBLAST/bin]$ ./mpiformatdb
Usage: ./mpiformatdb <--nfrags=<NumberOfFrags>|--frag-size=<SizeOfFrags> (in MB)> <formatdb arguments>
Options:
-f --config-file=<CONFIGFILE> Specify the location of the mpiBLAST configuration file
--nfrags=<NumberOfFrags> Specifies the number of fragments to split the database into
--frag-size=<SizeOfFrags> Specifies the size for database fragments, can not be used with --nfrags
--version Prints the version number
--debug Prints out debugging information
--enable-reorder Enable the sequences reordering..
--quiet Don't show % complete counter.
--update_db=<DBtoUpdate> Update database by adding additional fragments
Example:
mpiformatdb -i ecoli.nt --nfrags=5
Which divides the ecoli.nt database into 5 fragments.
Here is how the conversion is handled in both applications
function convertFastaTompiBLASTFasta (){
$mpiFormatDBExecutable -i $1 --nfrags=48 -pF
}
convertFastaTompiBLASTFasta $1
The mpiformatdb works with fasta databases, so either make sure to download the fasta version of the original or convert the original to the fasta format, which can be done using the tool fastacmd provided by the ncbi toolbox that came with mpiBLAST.
[login.budapest:/fs01/home/winger/mpiBLAST-1.6.0/ncbi/bin]$ ./fastacmd --help
fastacmd 2.2.20 arguments:
-d Database [String] Optional
default = nr
-p Type of file
G - guess mode (look for protein, then nucleotide)
T - protein
F - nucleotide [String] Optional
default = G
..
-D Dump the entire database as (default is not to dump anything):
1 FASTA
2 Gi list
3 Accession.version list
..
The code segment we use to convert the database
function convertDBtoFastaDB (){
$fastacmdExecutable -D 1 -d $1
if [ $? -eq 1 ]
then
errorHandler "011"
fi
mv $1 $1_original
mv "$1.fasta" $1
}
..
if [ "`checkDBType $1`" != "fasta" ]
then
convertDBtoFastaDB $1
fi
..
Using mpiBLAST
The mpiBLAST command line tool has almost exactly the same syntax as the original blastall program. The job can be executed using mpirun or mpiexec the following way:
mpirun -np $NSLOTS mpiBLAST -p blastn -d nt -i blast_query.fas -o blast_results.txt
Which starts mpiBLAST on $NSLOTS nodes, searches the nt database for the sequence found in blast_query.fas and stores the results in blast_results.txt (executing mpiBLAST locally from the console of a supercomputer would require the job to be submitted using qsub, but the ARC middleware used with the applications take care of that).
The code we use to execute mpiBLAST is the following
export mpiBLAST_SHARED=/scratch/hpsee/mpiBLAST/shared_temp /scratch/hpsee/mpiBLAST/bin/mpiBLAST $*
This script, when defined as an MPI job in the gUSE portal will be submitted to the supercomputer nodes, and the parameters for the job are passed to the mpiBLAST executable. This workaround is necessary because the current version of the gUSE portal (3.4) requires an executable to be uploaded when the job is defined - there is no direct way to specify a binary already present in the target system (as is the case with the mpiBLAST executable which has individually been compiled for all supercomputers with respect to the HW specification of each machine).
Usual input for mpiBLAST looks something like this
>test GCACCAGGGGACAGCGGCAACCATGAGTGACTCCACAGAAGCCAAGATGCAGCCTCTTAGCT CCATGGACGATGATGAGTTGATGGTCAGCGGCAGCAGGTATTCTATTAAAAGCTCCAGACTA CGACCAAATTCTGGAATCAAGTGTTTGGCAGGATGCTCGGGACACAGCCAAGTCCCCTTGGT CCTGCAGCTGCTCTCCTTCCTGTTCTTGGCTGGGCTCCTGCTGATCATTCTTTTCCAAGTCT CCAAAACCCCAAATACCGAGAGGCAGAAGGAACAAGAGAAGATCCTCCAGGAACTGACCCAG CTGACAGATGAGCTTACGTCCAGGATCCCCATCTCCCAAGGG
And the resulting file is
[login.budapest:/fs01/home/winger/shared_temp/DiseaseGeneMapper]$ vim results4.txt
gb|AW812589.1| CM4-ST0182-231199-049-g06 ST0182 Homo sapiens cDN... 44 0.72
gb|BG823875.1| 602729548F1 NIH_MGC_15 Homo sapiens cDNA clone IM... 42 2.9
...
emb|BX449065.2| BX449065 Homo sapiens FETAL LIVER Homo sapiens c... 42 2.9
>gb|CD673194.1| fg21h10.y1 Human Iris cDNA (Normalized): fg Homo sapiens cDNA clone
fg21h10 5', mRNA sequence
Length = 503
Score = 488 bits (246), Expect = e-134
Identities = 399/450 (88%)
Strand = Plus / Plus
Query: 653 ggacactttccaatggtgaggtgcagtgctgggcactgtggccggatgactcctactgga 712
||||||| ||||| ||||| |||||||||||||| |||||||| || |||||||||||||
Sbjct: 1 ggacactgtccaacggtgaagtgcagtgctgggccctgtggcctgacgactcctactgga 60
Query: 713 ccccgtacatgaccatcgtcgcctttctggtgtacttcattcccttggcaattatcagcg 772
|||| |||||||||||||| ||||| |||||||||||||| || || |||| ||||||
Sbjct: 61 ccccatacatgaccatcgtggccttcctggtgtacttcatccctctgacaatcatcagca 120
... (more follows)
mpiBLAST performance
We have originally set out to implement DeepAligner and DiseaseGeneMapper in BioPerl, but after the initial research realized that the performance offered by mpiBLAST would not be possible to achieve, so we have switched to using mpiBLAST instead, which offer an increase of performance of several orders of magnitude (http://www.mpiblast.org/About/Performance) compared to the standard NCBI BLAST tool. mpiBLAST performance has been covered and researched greatly over the years, a list of relevant publications can be found at the end of this section. According to [FENG 03] an 305x speedup is possible on 128 worker nodes, and [LIN 08] presents the result of their research where mpiBLAST achieved 93% efficiency on an IBM Blue Gene/P with 32.768 cores.
References
[LIN 08] Massively Parallel Genomic Sequence Search on the Blue Gene/P Architecture H. Lin, P. Balaji, R. Poole, C. Sosa, X. Ma, W. Feng IEEE/ACM SC2008: The International Conference on High-Performance Computing, Networking, and Storage, November 2008.
[FENG 03]: Green Destiny + mpiBLAST = Bioinfomagic W. Feng 10th International Conference on Parallel Computing (ParCo), September 2003. (6 pages - PDF, 203K)
for further information on mpiBLAST please refer to the mpiBLAST website
BLAS
Section contributed by IPB
BLAS (Basic Linear Algebra Subprograms) is a set of routines that provide standard building blocks for performing basic vector and matrix operations. The Level 1 BLAS routines perform scalar, vector and vector-vector operations, the Level 2 BLAS routines perform matrix-vector operations, and the Level 3 BLAS routines perform matrix-matrix operations. BLAS is commonly used in the development software that requires high quality linear algebra routines, as they are efficient, portable, and widely available. The reference BLAS is a freely-available software package and is available for download from the netlib web site (http://www.netlib.org/blas/).
Simple BLAS example using the C BLAS interface (http://www.netlib.org/blas/blast-forum/cblas.tgz) is illustrated with the code below. It uses DGEMV function to perform a simple matrix-vector multiplication. A corresponding C function from CBLAS is cblas_dgemv.
#include <stdio.h>
#include <cblas.h>
double m[] = {
3, 1, 3,
1, 5, 9,
2, 6, 5
};
double x[] = {
-1, -1, 1
};
double y[] = {
0, 0, 0
};
int main() {
int i, j;
for (i=0; i<3; ++i) {
for (j=0; j<3; ++j) printf("%5.1f", m[i*3+j]);
putchar('\n');
}
cblas_dgemv(CblasRowMajor, CblasNoTrans, 3, 3, 1.0, m, 3,
x, 1, 0.0, y, 1);
for (i=0; i<3; ++i) printf("%5.1f\n", y[i]);
return 0;
}
Machine-specific optimized BLAS libraries are available for a variety of computer architectures. Such libraries are provided by the computer vendors or by independent software vendors. Some popular optimized BLAS libraries are:
- AMD Core Math Library (ACML) - Mathematical library which provides mathematical routines optimized for AMD processors (including a full implementation of Level 1, 2 and 3 Basic Linear Algebra Subroutines).
- MKL - Intel's Math Kernel Library of optimized, math routines for science, engineering, and financial applications with math functions which include BLAS, LAPACK, ScaLAPACK, Sparse Solvers, Fast Fourier Transforms, and Vector Math.
- ESSL - IBM's Engineering and Scientific Subroutine Library, supporting the PowerPC architecture under AIX and Linux.
- libsci - The Cray LibSci library includes a number of parallel numerical libraries including ScaLAPACK, BLAS, BLACS, SuperLU and the Iterative Refinement Toolkit (IRT). The routines in LibSci can be called from both Fortran and C programs.
- Sun Performance Library – A library of optimized subroutines and functions for computational linear algebra and Fourier transforms. It is based on the standard libraries BLAS1, BLAS2, BLAS3, LINPACK, LAPACK, FFTPACK, and VFFTPACK.
Alternatively, a user can download ATLAS to automatically generate an optimized BLAS library for the target architecture. Some pre-built optimized BLAS libraries are also available from the ATLAS site. For a given set of machines, Goto BLAS implementation is also available.
FFTW
Section contributed by IPB
The Fastest Fourier Transform in the West (FFTW) is a comprehensive collection of C routines for computing the discrete Fourier transform (DFT) and its various special cases. Detailed description of FFTW features is available at the official web site (www.fftw.org) and on the HP-SEE wiki Software Stack and Technology Watch / Library (http://hpseewiki.ipb.ac.rs/index.php/Libraries).
Within the NUQG application, FFTW library is used for complex one-dimensional and multi-dimensional DFTs. It is used in serial, OpenMP and MPI parallel mode, as well as in the hybrid OpenMP/MPI mode. Here we describe NUQG FFTW serial and OpenMP usage in 1D, 2D, and 3D, while MPI and hybrid OpenMP/MPI FFTW implementation in NUQG is illustrated in the Hybrid programming and Distributed memory sections, respectively.
The following pseudo-codes show the serial usage of FFTW3 in 1D, 2D, and 3D codes. The codes have to be linked with the fftw3 library -lfftw3 -lm on Linux/Unix systems. In 1D DFT pseudo-code, Nx is the size of the transform. In multi-dimensional pseudo-codes, Nx and Ny define matrix size in 2D transforms, and Nx, Ny, Nz define tensor size in 3D transforms. FFTW_FORWARD and FFTW_BACKWARD tags in plan definitions indicate the direction of the transform. Once the plan has been created, it can be used as many times as needed for transforms on the specified input/output arrays, computing the actual transforms by calling the fftw_execute(plan) function. The plan is destroyed and the memory used de-allocated by calling the fftw_destroy_plan(plan) function. Computing the forward transform, followed by the backward transform (or vice versa) yields the original array multiplied by the size of the array Nx (in 1D case), size of the matrix Nx * Ny (in 2D case), or size of the tensor Nx * Ny * Nz (in 3D case), so it is necessary to rescale the data after each transformation.
// FFTW3 serial usage in 1D
#include <fftw3.h>
int main(int argc, char **argv) {
long cnti;
long Nx;
fftw_complex *array;
fftw_plan plan_forward, plan_backward;
array = ((fftw_complex *) fftw_malloc(sizeof(fftw_complex) * Nx));
plan_forward = fftw_plan_dft_1d(Nx, array, array, FFTW_FORWARD, FFTW_ESTIMATE);
plan_backward = fftw_plan_dft_1d(Nx, array, array, FFTW_BACKWARD, FFTW_ESTIMATE);
fftw_execute(plan_forward);
fftw_execute(plan_backward);
for(cnti = 0; cnti < Nx; cnti ++) {
array[cnti][0] /= Nx;
array[cnti][1] /= Nx;
}
fftw_destroy_plan(plan_forward);
fftw_destroy_plan(plan_backward);
fftw_free(array);
}
// FFTW3 serial usage in 2D
#include <fftw3.h>
int main(int argc, char **argv) {
long cnti;
long Nx, Ny;
fftw_complex *array;
fftw_plan plan_forward, plan_backward;
array = ((fftw_complex *) fftw_malloc(sizeof(fftw_complex) * Nx * Ny));
plan_forward = fftw_plan_dft_2d(Nx, Ny, array, array, FFTW_FORWARD, FFTW_ESTIMATE);
plan_backward = fftw_plan_dft_2d(Nx, Ny, array, array, FFTW_BACKWARD, FFTW_ESTIMATE);
fftw_execute(plan_forward);
fftw_execute(plan_backward);
for(cnti = 0; cnti < Nx * Ny; cnti ++) {
array[cnti][0] /= Nx * Ny;
array[cnti][1] /= Nx * Ny;
}
fftw_destroy_plan(plan_forward);
fftw_destroy_plan(plan_backward);
fftw_free(array);
}
// FFTW3 serial usage in 3D
#include <fftw3.h>
int main(int argc, char **argv) {
long cnti;
long Nx, Ny, Nz;
fftw_complex *array;
fftw_plan plan_forward, plan_backward;
array = ((fftw_complex *) fftw_malloc(sizeof(fftw_complex) * Nx * Ny * Nz));
plan_forward = fftw_plan_dft_3d(Nx, Ny, Nz, array, array, FFTW_FORWARD, FFTW_ESTIMATE);
plan_backward = fftw_plan_dft_3d(Nx, Ny, Nz, array, array, FFTW_BACKWARD, FFTW_ESTIMATE);
fftw_execute(plan_forward);
fftw_execute(plan_backward);
for(cnti = 0; cnti < Nx * Ny * Nz; cnti ++) {
array[cnti][0] /= Nx * Ny * Nz;
array[cnti][1] /= Nx * Ny * Nz;
}
fftw_destroy_plan(plan_forward);
fftw_destroy_plan(plan_backward);
fftw_free(array);
}
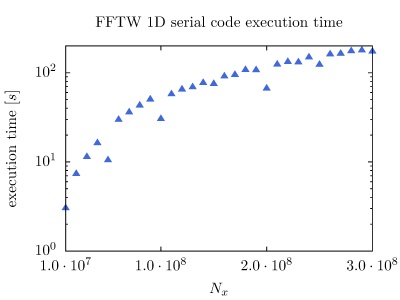
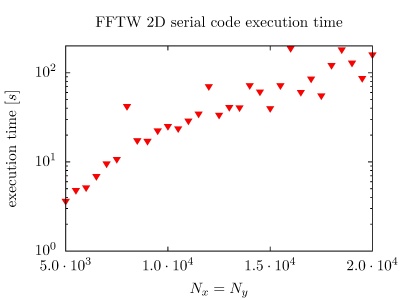
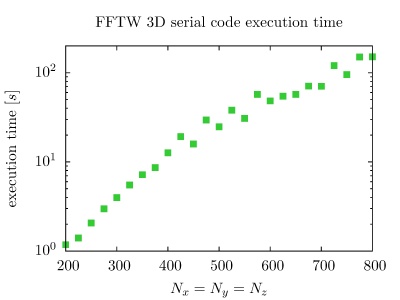
On the PARADOX cluster, the NUQG application uses FFTW library version 3.3.1. The execution times of the application in 1D, 2D, and 3D as a function of the array size, matrix size, and tensor size are given in the figures below.
The next set of pseudo-codes gives overview of the FFTW OpenMP usage in 1D, 2D, and 3D codes. Applications using the parallel complex transforms should be linked with the switches -lfftw3_threads -lfftw3 -lm, or -lfftw3_omp -lfftw3 -lm if the program is compiled with OpenMP. It is also needed to link against the library responsible for threads on your system (e.g. -lpthread on GNU/Linux), and to include a compiler flag that enables OpenMP (e.g. -fopenmp with GCC). Before calling any FFTW routines, the fftw_init_threads() function has to be invoked. This function, which needs to be called only once, performs initialization required for use of threads on the system. It returns zero if an error is encountered, and non-zero value otherwise.
Before creating FFTW3 plan for multi-threaded usage, is is necessary to call the fftw_plan_with_nthreads(int nthreads) function. The nthreads argument indicates the number of threads that will be used by FFTW. All plans created subsequently using any of the planner routines will use the same number of threads. Plans already created before a call to the function fftw_plan_with_nthreads are not affected. If nthreads is equal to 1 (default), threads are disabled for subsequent plans.
To configure FFTW to use all of the currently running OpenMP threads (set by the omp_set_num_threads(nthreads) function or by the OMP_NUM_THREADS environment variable), one should invoke fftw_plan_with_nthreads(omp_get_num_threads()). The 'omp_' OpenMP functions are declared via #include <omp.h> header file.
A given plan can be executed as before, by calling fftw_execute(plan), and the execution will use the number of threads specified when the plan was created. The plan can be destroyed the same as in the serial code, by calling fftw_destroy_plan, while the threads could be de-allocated by calling fftw_cleanup_threads(void).
// FFTW3 multi-threaded usage in 1D
#include <fftw3.h>
int main(int argc, char **argv) {
long cnti;
long Nx;
fftw_complex *array;
fftw_plan plan_forward, plan_backward;
array = ((fftw_complex *) fftw_malloc(sizeof(fftw_complex) * Nx));
fftw_init_threads();
fftw_plan_with_nthreads(omp_get_max_threads());
plan_forward = fftw_plan_dft_1d(Nx, array, array, FFTW_FORWARD, FFTW_ESTIMATE);
plan_backward = fftw_plan_dft_1d(Nx, array, array, FFTW_BACKWARD, FFTW_ESTIMATE);
fftw_execute(plan_forward);
fftw_execute(plan_backward);
for(cnti = 0; cnti < Nx; cnti ++) {
array[cnti][0] /= Nx;
array[cnti][1] /= Nx;
}
fftw_destroy_plan(plan_forward);
fftw_destroy_plan(plan_backward);
fftw_cleanup_threads();
fftw_free(array);
}
// FFTW3 multi-threaded usage in 2D
#include <fftw3.h>
int main(int argc, char **argv) {
long cnti;
long Nx, Ny;
fftw_complex *array;
fftw_plan plan_forward, plan_backward;
array = ((fftw_complex *) fftw_malloc(sizeof(fftw_complex) * Nx * Ny));
fftw_init_threads();
fftw_plan_with_nthreads(omp_get_max_threads());
plan_forward = fftw_plan_dft_2d(Nx, Ny, array, array, FFTW_FORWARD, FFTW_ESTIMATE);
plan_backward = fftw_plan_dft_2d(Nx, Ny, array, array, FFTW_BACKWARD, FFTW_ESTIMATE);
fftw_execute(plan_forward);
fftw_execute(plan_backward);
for(cnti = 0; cnti < Nx * Ny; cnti ++) {
array[cnti][0] /= Nx * Ny;
array[cnti][1] /= Nx * Ny;
}
fftw_destroy_plan(plan_forward);
fftw_destroy_plan(plan_backward);
fftw_cleanup_threads();
fftw_free(array);
}
// FFTW3 multi-threaded usage in 3D
#include <fftw3.h>
int main(int argc, char **argv) {
long cnti;
long Nx, Ny, Nz;
fftw_complex *array;
fftw_plan plan_forward, plan_backward;
array = ((fftw_complex *) fftw_malloc(sizeof(fftw_complex) * Nx * Ny * Nz));
fftw_init_threads();
fftw_plan_with_nthreads(omp_get_max_threads());
plan_forward = fftw_plan_dft_3d(Nx, Ny, Nz, array, array, FFTW_FORWARD, FFTW_ESTIMATE);
plan_backward = fftw_plan_dft_3d(Nx, Ny, Nz, array, array, FFTW_BACKWARD, FFTW_ESTIMATE);
fftw_execute(plan_forward);
fftw_execute(plan_backward);
for(cnti = 0; cnti < Nx * Ny * Nz; cnti ++) {
array[cnti][0] /= Nx * Ny * Nz;
array[cnti][1] /= Nx * Ny * Nz;
}
fftw_destroy_plan(plan_forward);
fftw_destroy_plan(plan_backward);
fftw_cleanup_threads();
fftw_free(array);
}
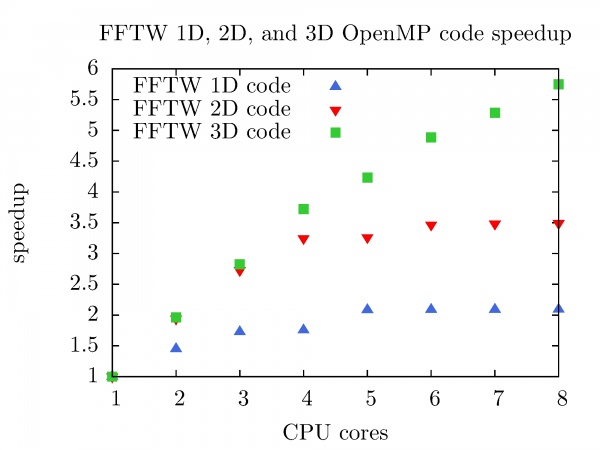
The speedup of FFTW OpenMP code in 1D, 2D, and 3D as a function of the number of CPU cores is shown in the figure below. FFTW 1D code (Nx = 3x10^8) on the PARADOX cluster with 8 CPU cores achieves speedup of 2.1. Better speedup is achieved with FFTW 2D code (Nx = Ny = 2 x 10^4) and FFTW 3D code (Nx = Ny = Nz = 800), 3.5 and 5.8 respectively.
It is worth mentioning that the speedup of FFTW 3D code with 8 CPU cores on the PARADOX cluster presented in this chapter is achieved by Intel MKL FFTW library already in for the case of 1D code (see section Mathematical and scientific libraries).
Intel MKL
Section contributed by IPB
Intel Math Kernel Library (Intel MKL) (http://software.intel.com/en-us/articles/intel-mkl/) is a library of optimized, extensively threaded mathematical routines for solving large scientific and engineering computational problems. The library supports C/C++ and Fortran language APIs. The most of Intel MKL functions are threaded, which enables more efficient usage of today’s multi-core processors and easier parallelization of application or implementation of a hybrid programming model. Threading is performed through OpenMP, and the following mathematical domains are supported:
- Sparse Linear Algebra – Sparse BLAS, Sparse Format Converters, PARDISO Direct Sparse Solver, Iterative Sparse Solvers and Pre-conditioners
- Fast Fourier Transforms
- Optimized LINPACK benchmark
- Vector Math Library
- Statistics Functions – Vector Random Number Generators, Summary Statistics Library
- Cluster Support – ScaLAPACK, Cluster FFT
The NUQG application uses Intel MKL Fast Fourier Transform and Trigonometric Transform functionality. The FFTW3 wrappers are a set of functions and data structures, depending on each other. The wrappers are not designed to provide the interface on a function-per-function basis, and some of the wrapper functions are empty and do nothing, but they are present to avoid linking errors and satisfy function calls.
The FFTW3 wrappers provide equivalent implementation for double- and single-precision functions (those with prefixes fftw_ and fftwf_, respectively). The FFTW3 interface provided by the wrappers is defined in the header files fftw3.h and fftw3.f. These files are identical to the ones from the FFTW3.2 package, and are included in the distribution of Intel MKL. Additionally, the files fftw3_mkl.h, fftw3_mkl.f, and fftw3_mkl_f77.h define supporting structures, supplementary constants and macros, and expose Fortran interface in C.
A function call for creation of an FFT plan may return a NULL plan, indicating that the functionality is not supported. Therefore, it is recommended to carefully check the result returned by a plan creation function within the application. In particular, the following problems return a NULL plan:
-
c2randr2cproblems with a split storage of complex data. -
r2rproblems with kind valuesFFTW_R2HC,FFTW_HC2R, andFFTW_DHT. - Multidimensional
r2rtransforms. - Transforms of multidimensional vectors.
- Multidimensional transforms with rank >
MKL_MAXRANK.
The new value MKL_RODFT00 is introduced for the kind parameter by the FFTW3 wrappers. For better performance, it is strongly suggested to use this value, rather than the FFTW_RODFT00:
plan1 = fftw_plan_r2r_1d(n, in1, out1, FFTW_RODFT00, FFTW_ESTIMATE); plan2 = fftw_plan_r2r_1d(n, in2, out2, MKL_RODFT00, FFTW_ESTIMATE);
The flags parameter in plan creation functions is always ignored. The same algorithm is used regardless of the value of this parameter. In particular, flags values FFTW_ESTIMATE, FFTW_MEASURE, etc. have no effect.
For multithreaded plans, the normal sequence of calls to the fftw_init_threads() and fftw_plan_with_nthreads() functions can be used.
FFTW3 wrappers are not fully thread-safe. If the new-array execute functions, such as fftw_execute_dft(), share the same plan from parallel user threads, we suggest to set the number of the sharing threads before creation of the plan. For this purpose, the FFTW3 wrappers provide a header file fftw3_mkl.h, which defines a global structure fftw3_mkl with a field to be set to the number of sharing threads. Below is an example of setting the number of sharing threads:
#include "fftw3.h" #include "fftw3_mkl.h" fftw3_mkl.number_of_user_threads = 4; plan = fftw_plan_dft(...);
The wrappers typically indicate a problem by returning a NULL plan. In few cases, the wrappers may report a descriptive message of the problem detected. By default, the reporting is turned off. To turn it on, one needs to set a variable fftw3_mkl.verbose to the non-zero value, for example:
#include "fftw3.h" #include "fftw3_mkl.h" fftw3_mkl.verbose = 0; plan = fftw_plan_r2r(...);
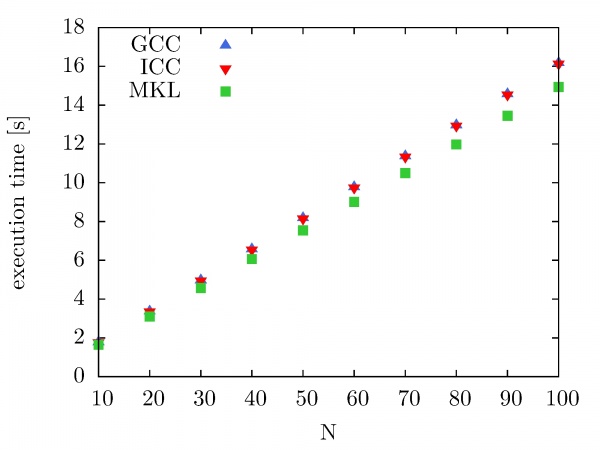
Intel MKL Fast Fourier Transform and Trigonometric Transform functionality is used by NUQG and tested on the PARADOX cluster. First, we have compared execution times of serial codes that use FFTW3 3.1.2 library compiled with the GCC compiler, compiled with the ICC compiler, as well as FFTW library provided by Intel MKL 10.2.3. The execution times are shown in the figure below as a function of a number of performed forward and backward DFT transforms. The size of the data array is 10^7. As we can see, there is no significant improvement in performance when the FFTW3 3.1.2 library is compiled with ICC instead of GCC. However, some improvement in performance is achieved when Intel MKL FFTW library is used.
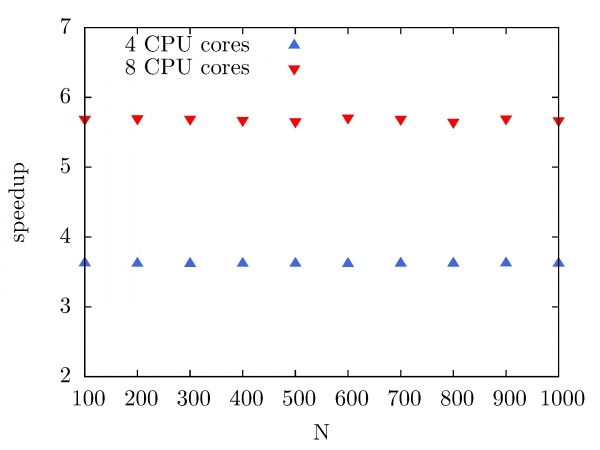
The next figure illustrates the speedup of the Intel MKL Fast Fourier Transform library on the PARADOX cluster as a function of the number of CPU cores used. In this case, the same data array size is used (10^7), while the number of performed forward and backward DFT transforms has been varied from 100 to 1000, with a step of 100. As we can see, the speedup of Intel MKL FFTW library on the PARADOX cluster is 3.8 for 4 CPU cores, and 5.8 for 8 CPU cores.
SPRNG
Section contributed by IPB
Pseudorandom number generation algorithms are key components in many modern scientific computing applications and the quality of the obtained research results often critically depends on their characteristics. The random number generating algorithms should be of sufficiently high quality, so as to ensure that numerical results give unbiased and reliable estimates of physical quantities. SPRNG (Scalable Parallel Random Number Generators) is one of the best libraries for generating pseudorandom numbers for parallel applications. Detail description of SPRNG features is available at the official web page (sprng.fsu.edu) and at the HP-SEE Wiki Software Stack and Technology Watch / Library (http://hpseewiki.ipb.ac.rs/index.php/Libraries) section.
Two versions of the SPRNG library are available on the PARADOX cluster: SPRNG 2.0 and SPRNG 4.0. The NUQG application uses SPRNG 4.0 library, both in the serial and in the MPI mode for the Monte Carlo calculation of path integrals of a generic quantum mechanical theory. Typical usage in the serial mode is illustrated with the following pseudo-code:
#include <sprng_cpp.h>
#define SEED 12345
int main()
{
int streamnum, nstreams;
double rn;
int irn;
Sprng * stream;
streamnum = 0;
nstreams = 1;
stream = SelectType(SPRNG_CMRG);
stream->init_sprng(streamnum, nstreams, SEED, SPRNG_DEFAULT);
// Generate a double precision random number
rn = stream->sprng();
// Generate an integer random number
irn = stream->isprng();
// Free memory used to store stream state
ptr->free_sprng();
}
The SPRNG MPI usage is illustrated with the following pseudo-code:
#include <sprng_cpp.h>
#define SEED 12345
int main(int argc, char *argv[])
{
int streamnum, nstreams;
Sprng *stream;
double rn;
int myid, nprocs;
// Initialize MPI
MPI_Init(&argc, &argv);
MPI_Comm_rank(MPI_COMM_WORLD, &myid);
MPI_Comm_size(MPI_COMM_WORLD, &nprocs);
streamnum = myid;
// One stream per processor
nstreams = nprocs;
//node 0 is reading in a generator type
stream = SelectType(SPRNG_CMRG);
// Initialize stream
stream->init_sprng(streamnum,nstreams,SEED,SPRNG_DEFAULT);
// Generate double precision random number
rn = stream->sprng();
// Free memory used to store stream state
stream->free_sprng();
// Terminate MPI
MPI_Finalize();
}
The NUQG Monte Carlo (MC) algorithm uses SPRNG streams independently, with almost no interprocess communication, and therefore the scaling of MPI NUQG MC algorithm with the number of processors is perfectly linear.
CPMD
Section contributed by IPB
The CPMD code is a parallelized plane-wave/pseudopotential implementation of the Density Functional Theory (DFT), particularly designed for ab-initio molecular dynamics. CPMD runs on a large variety of different computer architectures and is optimized to run on either (super-)scalar or vector CPUs. General description of CPMD and the list of features are available at the official web site (cpmd.org) and at the HP-SEE Wiki Software Stack and Technology Watch / Library section http://hpseewiki.ipb.ac.rs/index.php/CPMD.
CPMD is ported on the PARADOX cluster with MPI parallelization. The application is fully functional and can be started with the following command:
[dusan@ui ~]$ cpmd.x file.in [PP_path] > file.out
where file.in is an input file and [PP_path] is the location of pseudopotential files for all atomic species specified in the file.in input file.
The path to the pseudopotential library can be given in different ways:
- By setting the second command line argument
[PP_path]. - If the second command line argument
[PP_path]is not given, the program checks the environment variablesCPMD_PP_LIBRARY_PATHandPP_LIBRARY_PATH. - If neither the environment variables nor the second command line argument are set, the program assumes that the pseudopotential files reside in the current directory.
During the run, cpmd.x creates different outputs:
- Various status messages to monitor the correct operation of the program is printed to standard output (in our case, redirected to the file
file.out).
- Detailed data are written into different files (depending on the keywords specified in the input
file.in). Large files are written either to the current directory, the directory specified by the environment variableCPMD_FILEPATH, or the directory specified in the input file using the keywordFILEPATH.
The CPMD official web site provides several CPMD introduction guides:
- Wavefunction Optimization Example - demonstrate some of the basic steps of performing a CPMD calculation with a very simple molecule: hydrogen, and a very simple task: calculate the electronic structure.
http://cpmd.org/cpmd-tutorial/the-basics-running-cpmd-input-and-output-formats
- Geometry Optimization and MD Example - inspect how to perform a geometry optimization and how to setup and analyze a Car-Parrinello MD.
http://cpmd.org/cpmd-tutorial/the-basics-of-cpmd-geometry-optimization-and-md
- Electron Structure and Geometry Optimization Example - gives three exercises (Hydrogen molecule, Water and Ammonia) to practice with the basics of CPMD.
http://cpmd.org/cpmd-tutorial/exercise-1-electron-structure-and-geometry-optimization
- Car-Parrinello MD Example - explain how to setup a Car-Parrinello Molecular Dynamics run.
http://cpmd.org/cpmd-tutorial/exercise-2-car-parrinello-md
- Bulk Systems Example - look into more complex systems like bulk materials.
http://cpmd.org/cpmd-tutorial/exercise-3-bulk-systems
- Determination of Dynamical Properties Example - is a collection of some examples on how to determine and tune dynamical properties in CPMD.
http://cpmd.org/cpmd-tutorial/exercise-4-determination-of-dynamical-properties
- Proton transfer in an Enzyme Example - shows how to setup/run a model system for a real-life simulation.
http://cpmd.org/cpmd-tutorial/exercise-5-triade
GAMESS
Section contributed by IICT-BAS
The GAMESS application was used by the MD Cisplatin application. In order to explore the capabilities of the free quantum-chemical software Firefly PCGamess, appropriate purely organic molecules were chosen, with the aid, on a later stage the program to be applied to metal-containing systems, like cisplatin analogues. The work done has thus an important methodological significance. Molecular mechanics conformational search for the two species was performed, and the global minimum-energy conformations thus obtained, were further optimized at HF ab initio (3-21G** basis set, Firefly PCGamess) level of theory. In both cases, specific, scorpion-like conformations are realized, with hydrogen bonds involving the guanidino-group and the phenolic hydroxyl. The application was porting to MPI using GAMESS application software on the HPCG cluster at IICT-BAS. A lot of MPI tests were done up to 32 cores to optimize the application. A good parallel efficiency is obtained up to 32 cores on HPCG cluster. Some problems were solved by testing different versions of Gamess software application and selecting the version that achieves correct execution. This may be due to some over-optimisation by the compiler. The GAMESS application is rather flexible and during configuration phase one can select different standard libraries and compiler flags. All tasks using Gamess were compiled on the HPCG cluster using Intel Math Kernel Library, which resulted in good overall performance.
libtiff
Section contributed by UVT
FuzzyCmeans used a special library, libtiff, to deal with the digital images involved in the computational process. Libtiff (http://www.libtiff.org/) was compiled for BG/P@UVT and InfraGRID cluster without any specific optimizations.
The workflow dealing with satellite images (most of the time in TIFF format) using libtiff library is the following:
1. open the TIFF image in memory
TIFF* image;
if((image=TIFFOpen(name , "w")) == NULL)
{
printf("Error open the output file");
return -1;
}
2. read image header and set some global variables for later use (image coordinates, image size, image bands etc.)
TIFFGetField(image, TIFFTAG_BITSPERSAMPLE, BPS); TIFFGetField(image, TIFFTAG_SAMPLESPERPIXEL, SPP); TIFFGetField(image, TIFFTAG_ROWSPERSTRIP, RPS); TIFFGetField(image, TIFFTAG_XRESOLUTION, XR); TIFFGetField(image, TIFFTAG_YRESOLUTION, YR); TIFFGetField(image, TIFFTAG_RESOLUTIONUNIT, RuN); TIFFGetField(image,TIFFTAG_IMAGEWIDTH, width); TIFFGetField(image,TIFFTAG_IMAGELENGTH, height);
3. read image values (define a structure that can hold the entire image, read each pixel value and assign it to the tiff structure)
uint32 *raster= (uint32*) _TIFFmalloc(width*height*sizeof(uint32));
// read the entire image
if(TIFFReadRGBAImageOriented(image, width, height, raster, ORIENTATION_TOPLEFT, 1))
{
// process the raster to compute the image
}
4. when all computations are done then write the virtual image from memory on the disk and free up memory
char *buffer;
TIFF* image;
buffer=malloc(width*height*sizeof(char));
// open a new file on disk for writting
if((image=TIFFOpen(name , "w")) == NULL)
{
printf("Error open the output file");
return -1;
}
// set initial TIFF headers
TIFFSetField(image,TIFFTAG_IMAGEWIDTH, width);
TIFFSetField(image,TIFFTAG_IMAGELENGTH, height);
TIFFSetField(image, TIFFTAG_BITSPERSAMPLE, 8);
TIFFSetField(image, TIFFTAG_SAMPLESPERPIXEL, 1);
TIFFSetField(image, TIFFTAG_ROWSPERSTRIP, TIFFDefaultStripSize(image, 0));
TIFFSetField(image, TIFFTAG_PHOTOMETRIC, PHOTOMETRIC_MINISWHITE);
TIFFSetField(image, TIFFTAG_FILLORDER, FILLORDER_MSB2LSB);
TIFFSetField(image, TIFFTAG_PLANARCONFIG, PLANARCONFIG_CONTIG);
TIFFSetField(image, TIFFTAG_XRESOLUTION, XR);
TIFFSetField(image, TIFFTAG_YRESOLUTION, YR);
TIFFSetField(image, TIFFTAG_RESOLUTIONUNIT, RuN);
// For each pixel find the closest center and set the corresponding gray level in the output image
for(i=0;i<height*width;i++) {
// fill the buffer with gray level values
…
Buffer[i] = value;
…
}
// write values to the file
for(i=0;i<height;i++)
{
TIFFWriteScanline(image,&buffer[i*width], i,0);
}
TIFFClose(image);
The snippets presented are only parts of the entire program and must be used as an usage example. For more details please consult the library API.
VMD
Section contributed by IFIN-HH
The Visual Molecular Dynamics (VMD) program provides 3-D visualization of large biomolecular systems and means for their modeling and analysis.
In the HP-SEE ecosystem, VMD is currently used by the ISyMAB application, which is hosted on the IFIN_Bio cluster, for the post- processing of the data obtained through the modeling of large biomolecular systems by means of the parallel molecular dynamics NAMD code.
The ISyMAB application provides access to a remote VMD server, hosted on the IFIN_Bio graphic server. In order to get access, the user must be authenticated on the ISyMAB web server through username / password or SSL certificate.
After authentication, one must initiate a session with the VMD graphic server. For this, one must click on the Manage Servers icon, and then on VNC. Depending on the available bandwidth and the quality of the Internet connection, one must select from the drop-down menu one of the three connection types:
- High Quality (more than 50Mbps),
- Medium Quality (2-50 Mbps),
- Low Quality (less than 2Mbps),
and then hit the GO button.
The next figure shows a link to the graphic server which runs VMD. In the new window, the user has a launcher of the VMD.
In order to have a good user experience, a maximum number of 3 simultaneous VMD user connections is currently allowed. Also, when the user logs out, the VMD session will be automatically closed in order to let other users get in.
OpenCV
Section contributed by UPB
Computer vision is everywhere - in security systems, manufacturing inspection systems, medical image analysis, Unmanned Aerial Vehicles, and more. It stitches Google maps and Google Earth together, checks the pixels on LCD screens, and makes sure the stitches in your shirt are sewn properly. OpenCV provides an easy-to-use computer vision framework and a comprehensive library with more than 500 functions that can run vision code in real time. Thus OpenCV (Open Source Computer Vision) is a library of programming functions for real time computer vision. OpenCV is written in C/C++ and Python and more recently supports GPUs. More specifically, the OpenCV GPU module is a set of classes and functions that utilize GPU computational capabilities. It is implemented using NVidia CUDA Runtime API, so only that vendor GPUs are supported. It includes utility functions, low level vision primitives as well as high level algorithms. The module is being developed as power infrastructure for fast vision algorithms building on GPU with some high level state of the art functionality. The CPU version of the library is employs a TBB parallelization strategy.
Here is an example of OpenCV usage within the EagleEye Project - in a function that processes satellite images:
void process_satellite_file(char* file_name, thread_data* tdp) {
/*process a single file*/
/*OpenCV Image structure pointers for original image, blurred image,
black and white image, image obtained after canny and image obtained after
histogram equalization */
IplImage *img_orig, *img_blurred, *img_gray, *img_canny, *img_equal = NULL;
CvMemStorage* storage = cvCreateMemStorage(0);
/*some floats used for printing time statistics at the end*/
float time1 = 0.0, time2 = 0.0, time3 = 0.0, time4 = 0.0, time5 = 0.0,time6 = 0.0;
/*timeval structures used for time statistics*/
struct timeval start, end;
/*some char* for output file_name generation*/
char buffer[MAX_FILE_NAME_SIZE], buffer2[MAX_FILE_NAME_SIZE];
char* pch = NULL;
config_data* cdp = &tdp->cd;
segment_data* sdp = &tdp->sd;
int class_res;
printf("[%d,%ld] Processing: %s\n", tdp->mpi_rank, tdp->tid, file_name);
/*load original image from file*/
img_orig = cvLoadImage(file_name, CV_LOAD_IMAGE_COLOR);
if (img_orig == NULL) {
fprintf(stderr, "%s:%d: Can't read image %s\n", __func__, __LINE__, file_name);
return;
}
class_res = classify(file_name, img_orig, cdp);
if (class_res != 42) {
cvReleaseImage(&img_orig);
return;
}
/*initializes image structures*/
img_blurred = cvCreateImage(cvSize(cdp->chunk_size, cdp->chunk_size), img_orig->depth, img_orig->nChannels);
img_gray = cvCreateImage(cvSize(cdp->chunk_size, cdp->chunk_size), img_orig->depth, 1);
img_canny = cvCreateImage(cvSize(cdp->chunk_size, cdp->chunk_size), img_orig->depth, 1);
int _height = img_orig->height - img_orig->height % cdp->chunk_size;
int _width = img_orig->width - img_orig->width % cdp->chunk_size;
/*if color matching is used load the road samples*/
if (cdp->use_color_match && tdp->nr_pixels1 == -1) {
load_road_samples(cdp, tdp, SATELLITE_MODE);
}
/*if histogram equalization is used*/
if (cdp->use_hist_equal) {
img_equal = cvCreateImage(cvSize(cdp->chunk_size, cdp->chunk_size), img_orig->depth, 1);
}
/*main processing loop - split image into square chunks and process each chunk*/
for (int y = 0; y < _height; y += cdp->chunk_size)
for (int x = 0; x < _width; x += cdp->chunk_size) {
/*set region of interest to selected chunk*/
CvRect rect = cvRect(x, y, cdp->chunk_size, cdp->chunk_size);
cvSetImageROI(img_orig, rect);
/* median + grayscale conversion + hist equalization + canny + hough
for different sizes of the median filter*/
for (int k = cdp->median_min_filter_size;
k <= cdp->median_max_filter_size;
k += 2) {
//median filter (blur)
gettimeofday(&start, NULL);
median_filter(img_orig, img_blurred, k, y, x);
gettimeofday(&end, NULL);
time1 += GET_TIME(start, end);
//grayscale conversion
gettimeofday(&start, NULL);
convert_to_grayscale(img_blurred, img_gray, y, x);
gettimeofday(&end, NULL);
time4 += GET_TIME(start, end);
//histogram equalization
if (cdp->use_hist_equal) {
gettimeofday(&start, NULL);
hist_equal(img_gray, img_equal);
gettimeofday(&end, NULL);
time6 += GET_TIME(start, end);
}
//canny filter
gettimeofday(&start, NULL);
if (cdp->use_hist_equal)
adaptive_canny(img_equal, img_canny, y, x,
cdp->canny_max_tries, cdp);
else
adaptive_canny(img_gray, img_canny, y, x,
cdp->canny_max_tries, cdp);
gettimeofday(&end, NULL);
time2 += GET_TIME(start, end);
//hough transform
int k2 = HOUGH_GAP;
gettimeofday(&start, NULL);
hough(img_canny, storage, x, y, k * 2, k2, cdp, sdp);
gettimeofday(&end, NULL);
time3 += GET_TIME(start, end);
}
}
//deactivate region of interest
cvResetImageROI(img_orig);
/*filter chosen segments if color matching is used*/
if (cdp->use_color_match) {
gettimeofday(&start, NULL);
color_filter_segments(img_orig, cdp, sdp, tdp->pixels1, tdp->nr_pixels1);
gettimeofday(&end, NULL);
time5 = GET_TIME(start, end);
}
//draw the segments over the original image and write the image to a file
draw_segments(img_orig, cdp, sdp);
for (int k = strlen(file_name) - 1; k >= 0; k--)
if (file_name[k] == '/') {
pch = &file_name[k + 1];
break;
}
memset(buffer, 0, MAX_FILE_NAME_SIZE);
memset(buffer2, 0, MAX_FILE_NAME_SIZE);
strncpy(buffer, pch, strlen(pch) - 4); //assuming all files end in .jpg
strcat(buffer, "_sat.jpg");
strcpy(buffer2, tdp->output_folder);
strcat(buffer2, "/");
strcat(buffer2, buffer);
cvSaveImage(buffer2, img_orig);
//free all the OpenCV images
if (cdp->use_hist_equal)
cvReleaseImage(&img_equal);
cvReleaseImage(&img_blurred);
cvReleaseImage(&img_canny);
cvReleaseImage(&img_gray);
cvReleaseImage(&img_orig);
//print time statistics
printf("\n[%d,%ld] Processed: %s\n"
"Time (seconds):\n\tmedian %lf\n\tconvert_gray %lf\n\thist_equal %lf"
"\n\tcanny %lf\n\though %lf\n\tcolor_filter_segments %lf\n\t"
"total %lf\n\n", tdp->mpi_rank, tdp->tid, file_name,
time1, time4, time6, time2, time3, time5,
time1 + time2 + time3 + time4 + time5 + time6);
fflush(stdout);
}
Within the HP-SEE virtual research communities, the OpenCV library is used by the UPB team in the development of the EagleEye project, implementing Feature Extraction from Satellite Images Using Hybrid Computing Architectures – CPU-GPU. Also we can envisage a joint usage of OpenCV features with the Computer Science Department from the West University of Timisoara, and the Romanian Institute for Space Sciences for satellite image processing and manipulation.
The current license for the OpenCV library is the BSD license.
Relevant and up-to-date resources are:
http://sourceforge.net/projects/opencv/
http://opencv.willowgarage.com/wiki/
ScaLAPACK
Section contributed by UPB
ScaLAPACK is a library of high-performance linear algebra routines for distributed-memory message-passing MIMD computers and networks of workstations supporting PVM or MPI. It is a continuation of the LAPACK project, which designed and produced analogous software for workstations, vector supercomputers, and shared-memory parallel computers. Both libraries contain routines for solving systems of linear equations, least squares problems, and eigenvalue problems. The goals of both projects are efficiency (to run as fast as possible), scalability (as the problem size and number of processors grow), reliability (including error bounds), portability (across all important parallel machines), flexibility (so users can construct new routines from well-designed parts), and ease of use (by making the interface to LAPACK and ScaLAPACK look as similar as possible). Many of these goals, particularly portability, are aided by developing and promoting standards, especially for low-level communication and computation routines. We have been successful in attaining these goals, limiting most machine dependencies to two standard libraries called the BLAS, or Basic Linear Algebra Subprograms, and BLACS, or Basic Linear Algebra Communication Subprograms. LAPACK will run on any machine where the BLAS are available, and ScaLAPACK will run on any machine where both the BLAS and the BLACS are available. Like LAPACK, the ScaLAPACK routines are based on block-partitioned algorithms in order to minimize the frequency of data movement between different levels of the memory hierarchy. The fundamental building blocks of the ScaLAPACK library are distributed memory versions (PBLAS) of the Level 1, 2 and 3 BLAS, and a set of Basic Linear Algebra Communication Subprograms (BLACS) for communication tasks that arise frequently in parallel linear algebra computations.
The library is currently written in Fortran 77 (with the exception of a few symmetric eigenproblem auxiliary routines written in C to exploit IEEE arithmetic) in a Single Program Multiple Data (SPMD) style using explicit message passing for interprocessor communication. The name ScaLAPACK is an acronym for Scalable Linear Algebra PACKage, or Scalable LAPACK. ScaLAPACK is designed for heterogeneous computing and is portable on any computer that supports MPI or PVM.
Within the HP-SEE virtual research communities, the ScaLAPACK library can be used by all partners that make use of the LAPACK library. In particular, the University Politehnica of Bucharest, the Institute of Chemistry from the Faculty of Natural Science and Mathematics in Macedonia, the NIIFI Supercomputing Center in Hungary, and the Institute of Physics Belgrade Serbia.
The current license for the OpenCV library is the BSD-new license.
Relevant resources can be found here: