Adapting code to memory architecture
From HP-SEE Wiki
(→HPC clusters) |
(→HPC clusters) |
||
| Line 157: | Line 157: | ||
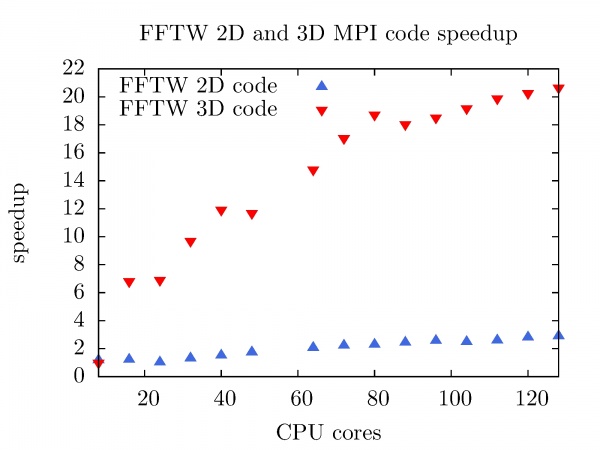
The speedup of FFTW MPI code in 2D, and 3D as a function of the number of CPU cores at PARADOX cluster is shown in the figure below. Better and useful speedup is achieved with FFTW 3D code (Nx = Ny = Nz = 1000), while the FFTW 2D MPI code for the small matrix (Nx = Ny = 2 x 10^4) is totally useless in the sense of the performance. | The speedup of FFTW MPI code in 2D, and 3D as a function of the number of CPU cores at PARADOX cluster is shown in the figure below. Better and useful speedup is achieved with FFTW 3D code (Nx = Ny = Nz = 1000), while the FFTW 2D MPI code for the small matrix (Nx = Ny = 2 x 10^4) is totally useless in the sense of the performance. | ||
| + | |||
| + | <center> | ||
| + | [[File:Fftw-mpi.jpg|600px]] | ||
| + | </center> | ||
=== BG/P === | === BG/P === | ||
Latest revision as of 18:27, 18 April 2012
Contents |
Symmetric multiprocessing
SMP on BG/P
Section contributed by IICT-BAS
Each Blue Gene/P Compute chip contains four PowerPC 450 processor cores, running at 850 MHz. The cores are cache coherent and the chip can operate as a 4-way symmetric multiprocessor (SMP). The memory subsystem on the chip consists of small private L2 caches, a central shared 8 MB L3 cache, and dual DDR2 memory controllers. The bottleneck in the scalability of SMP using crossbar switches is the bandwidth and power consumption of the interconnect among the various processors, the memory, and the disk arrays. For SMP Model in BlueGene/P, each node performs a copy of the program and it can run 1-4 threads, each running of the processor within the node. One should be aware that on the Blue Gene there is no virtual memory. Memory is not divided between CPU cores and thus SMP programs on one CPU are possible, however at most 4 threads can be run. Thus it is more logical for HPC usage to use a hybrid OpenMP - MPI computation model. In such a model there is additional advantage in that the number of MPI processes is less than a pure MPI program would require and thus communication is simplified and less communication buffers are required. A Blue Gene node may be run in three modes - SMP,DUAL,VN. In SMP mode there is one MPI process per node, 4GB memory, up to 4 threads. In DUAL mode there are two MPI processes per node, 2GB memory per process, up to two threads. In virtual-node mode (VN) there are four MPI processes per node, 1GB limit per process.
ccNUMA
Section contributed by NIIFI
Non-Uniform Memory Access (NUMA) is a computer memory design used in Multiprocessing, where the memory access time depends on the memory location relative to a processor. Under NUMA, a processor can access its own local memory faster than non-local memory, that is, memory local to another processor or memory shared between processors. Nearly all CPU architectures use a small amount of very fast non-shared memory known as cache to exploit locality of reference in memory accesses. With NUMA, maintaining cache coherence across shared memory has a significant overhead. NUMA computers are using special-purpose hardware to maintain cache coherence, and thus class as "cache-coherent NUMA", or ccNUMA. Typically, this takes place by using inter-processor communication between cache controllers to keep a consistent memory image when more than one cache stores the same memory location. For this reason, ccNUMA may perform poorly when multiple processors attempt to access the same memory area in rapid succession. Because the Linux operating system has a tendency to migrate processes, the importance of using a placement tool becomes more apparent.
Useful placement tools on the SGI UV machine
dplace
You can use the dplace command to bind a related set of processes to specific CPUs or nodes to prevent process migration. By default, memory is allocated to a process on the node on which the process is executing. If a process moves from node to node while it running, a higher percentage of memory references are made to remote nodes. Because remote accesses typically have higher access times, process performance can be diminished. CPU instruction pipelines also have to be reloaded.
Using the dplace command with MPI Programs:
mpirun -np 12 /usr/bin/dplace -s1 -c 24-35 ./connectivity
Using dplace command with OpenMP Programs:
export OMP_NUM_THREADS 6 /usr/bin/dplace -x6 –c 24-29 ./program</nowiki>
numactl
It runs processes with a specific NUMA scheduling or memory placement policy. The policy is set for command and inherited by all of its children. The processes will be bind to specific memory nodes in this case. Example for numactl:
/usr/bin/numactl –m 5 ./program
The program can access only the number 5 memory node.
cpuset
The cpuset file system is a pseudo-file-system interface to the kernel cpuset mechanism, which is used to control the processor placement and memory placement of processes. It is commonly mounted at /dev/cpuset. Automatic cpuset allocation has been configured on the Pecs UV [pecs] machine which is integrated to the Sun Grid Engine (SGE) scheduler. If a new job is starting then the SGE create a new cpuset for the job which contain only the asked CPUs and their local memory nodes. Other jobs cannot access these.
RESOURCES
[pecs] http://wiki.hp-see.eu/index.php/Resource_centre_Pecs_SC Pecs UV
Distributed memory
HPC clusters
Section contributed by IPB
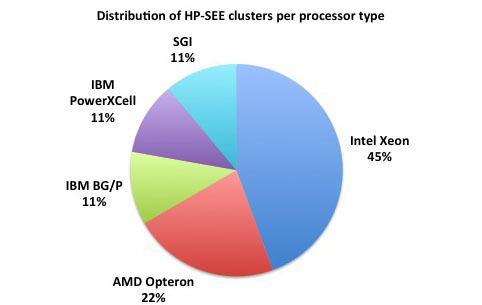
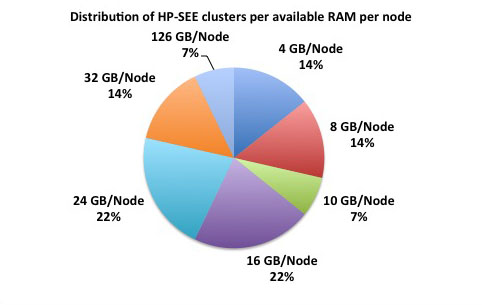
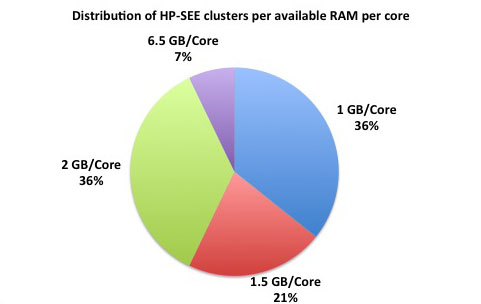
Distributed memory refers to a multiple-processor computer system in which each processor has its own private memory. Computational tasks can only operate on local data, and if remote data is required, the computational task must communicate with one or more remote processors. As it is illustrated at the first figure below, 78% of the HP-SEE resource centers are HPC distributed memory systems. 45% of total number of HP-SEE clusters are Intel Xeon based distributed memory systems, while 22% are AMD Opteron based, and in 11% it is IBM BG/P systems. The second figure gives distribution of HP-SEE clusters per available RAM per node, and third figure shows distribution of HP-SEE clusters per available RAM per core. In most of the cases, HP-SEE distributed memory systems have 16, 24, or 32 GB/Node, which results with 1.5, or 2 GB/Core.
Usage of HPC distributed memory resource centers requires distributed-memory parallelism. This kind of parallelism is especially useful in transformation of arrays so large that they do not fit into the memory of a single processor. The storage per-process required by this parallelism is proportional to the total array size divided by the number of processes. Conversely, distributed-memory parallelism can easily pose an unacceptably high communications overhead for small problems. The threshold problem size for which parallelism becomes advantageous depends on the precise problem, available hardware (interconnection), and application implementation.
MPI is a language-independent communications protocol used to program distributed-memory HPC systems. Most MPI implementations consist of a specific set of routines (i.e., an API) directly callable from C, C++, Fortran and any language able to interface with such libraries, including C#, Java or Python. The advantages of MPI over older message passing libraries are portability (because MPI has been implemented for almost every distributed memory architecture) and speed (because each implementation is in principle optimized for the hardware on which it runs).
The usage of HPC distributed memory resource centers by MPI parallelized application here will be demonstrated by NUQG application. While the NUQG FFTW serial and OpenMP implementation in 1D, 2D, and 3D is illustrated Libraries usage / FFTW section, the following pseudo-codes show the MPI FFTW3 library usage in NUQG:
// FFTW3 MPI usage in 2D
#include <fftw3-mpi.h>
int main(int argc, char **argv) {
int nproc, procid;
long cnti;
long seed, iter, Nx, Ny;
ptrdiff_t larray, lNx, lstart;
fftw_complex *array;
fftw_plan plan_forward, plan_backward;
MPI_Init(&argc, &argv);
MPI_Comm_size(MPI_COMM_WORLD, &nproc);
MPI_Comm_rank(MPI_COMM_WORLD, &procid);
fftw_mpi_init();
larray = fftw_mpi_local_size_2d(Nx, Ny, MPI_COMM_WORLD, &lNx, &lstart);
array = fftw_alloc_complex(larray);
plan_forward = fftw_mpi_plan_dft_2d(Nx, Ny, array, array, MPI_COMM_WORLD, FFTW_FORWARD, FFTW_ESTIMATE);
plan_backward = fftw_mpi_plan_dft_2d(Nx, Ny, array, array, MPI_COMM_WORLD, FFTW_BACKWARD, FFTW_ESTIMATE);
fftw_execute(plan_forward);
fftw_execute(plan_backward);
for(cnti = 0; cnti < lNx * Ny; cnti ++) {
array[cnti][0] /= Nx * Ny;
array[cnti][1] /= Nx * Ny;
}
fftw_destroy_plan(plan_forward);
fftw_destroy_plan(plan_backward);
fftw_free(array);
fftw_mpi_cleanup();
MPI_Finalize();
}
// FFTW3 MPI usage in 3D
#include <fftw3-mpi.h>
int main(int argc, char **argv) {
int nproc, procid;
long cnti, cntj;
long seed, iter, Nx, Ny, Nz;
double exetime, errtime, tmptime;
ptrdiff_t larray, lNx, lstart;
fftw_complex *array;
fftw_plan plan_forward, plan_backward;
MPI_Init(&argc, &argv);
MPI_Comm_size(MPI_COMM_WORLD, &nproc);
MPI_Comm_rank(MPI_COMM_WORLD, &procid);
fftw_mpi_init();
larray = fftw_mpi_local_size_3d(Nx, Ny, Nz, MPI_COMM_WORLD, &lNx, &lstart);
array = fftw_alloc_complex(larray);
plan_forward = fftw_mpi_plan_dft_3d(Nx, Ny, Nz, array, array, MPI_COMM_WORLD, FFTW_FORWARD, FFTW_ESTIMATE);
plan_backward = fftw_mpi_plan_dft_3d(Nx, Ny, Nz, array, array, MPI_COMM_WORLD, FFTW_BACKWARD, FFTW_ESTIMATE);
fftw_execute(plan_forward);
fftw_execute(plan_backward);
for(cnti = 0; cnti < lNx * Ny * Nz; cnti ++) {
array[cnti][0] /= Nx * Ny * Nz;
array[cnti][1] /= Nx * Ny * Nz;
}
fftw_destroy_plan(plan_forward);
fftw_destroy_plan(plan_backward);
fftw_free(array);
fftw_mpi_cleanup();
MPI_Finalize();
}
MPI FFTW routines should be linked with -lfftw3_mpi -lfftw3 -lm on Unix in double precision. In addition, it is necessary to link whatever library is responsible for MPI on the system. In most MPI implementations, there is a special compiler alias named mpicc to compile and link MPI code.
After calling MPI_Init, call of FFTW fftw_mpi_init() function is required. With function fftw_mpi_plan_dft_2d plan is created, while the standard fftw_execute and fftw_destroy routines are used for the MPI transform. As it is illustrated in the code above, the FFTW MPI routines take ptrdiff_t arguments instead of int as for the serial FFTW. code>ptrdiff_t</code> is a standard C integer type which is (at least) 32 bits wide on a 32-bit machine and 64 bits wide on a 64-bit machine.
FFTW MPI doesn't allocate the entire 2D/3D array on each process. Instead, fftw_mpi_local_size_2d/fftw_mpi_local_size_3d functions find out what portion of the array resides on each processor, and how much space to allocate. Here, the portion of the array on each process is a lNx by Ny (Ny * Nz in 3D) slice of the total array, starting at index lstart. The total number of fftw_complex numbers to allocate is given by the larray return value, which may be greater than larray * Ny (larray * Ny * Nz in 3D).
The speedup of FFTW MPI code in 2D, and 3D as a function of the number of CPU cores at PARADOX cluster is shown in the figure below. Better and useful speedup is achieved with FFTW 3D code (Nx = Ny = Nz = 1000), while the FFTW 2D MPI code for the small matrix (Nx = Ny = 2 x 10^4) is totally useless in the sense of the performance.
BG/P
Section contributed by IICT-BAS
As an example application that is adapting the code to memory architecture on IBM BlueGene/P we consider the Gromacs software package, which includes Global Array Toolkit library. Global Array Toolkit is deployed and can be used by user codes. It is an efficient platform-independent API, which provides the so-called Global Array abstraction. This is an array, whose storage is distributed in the memory of all MPI processes, but from the application point of view is accessed uniformly, as if it were in the local memory. This abstracts and hides the distributed nature of the memory of the host machine and one can create arrays as large as needed, as long as they fit in the total combined memory of all computing nodes. The Global array toolkit has additional libraries, including a Memory Allocator (MA), Aggregate Remote Memory Copy Interface (ARMCI), and functionality for out-of-core storage of arrays (ChemIO). Shared memory is used to store global array data and is allocated by the Global Arrays run-time system called ARMCI. ARMCI uses shared memory to optimize performance and avoid explicit interprocessor communication within a single shared memory system or an SMP node. ARMCI allocates shared memory from the operating system in large segments and then manages memory in each segment in response to the GA allocation and deallocation calls. Each segment can hold data in many small global arrays. ARMCI does not return shared memory segments to the operating system until the program terminates. Another application that can be compiled with ARMCI support is GAMESS.