CMSLTM
From HP-SEE Wiki
(→Running on Several HP-SEE Centres) |
(→Running on Several HP-SEE Centres) |
||
| Line 100: | Line 100: | ||
== Running on Several HP-SEE Centres == | == Running on Several HP-SEE Centres == | ||
| - | [[CMSLTM/Scalability Click here for full Benchmark Analysis]] | + | [[CMSLTM/Scalability|Click here for full Benchmark Analysis]] |
Latest revision as of 14:22, 16 January 2013
General Information
- Application's name: Computational Models of Short and Long Term Memory
- Application's acronym: CMSLTM
- Virtual Research Community: Life Sciences
- Scientific contact: Dr. Panayiota Poirazi, poirazi@imbb.forth.gr
- Technical contact: George Kastellakis, gkastel@imbb.forth.gr
- Developers: George Kastellakis, IMBB-FORTH, Greece
- Web site: http://www.imbb.forth.gr/people/poirazi/drupal/?q=node/7
Short Description
This project involves the development of biologically relevant compartmentalized models of neurons and neuronal networks. We are interested in the modeling of processes related to:
1. Models of sustained activity in the Prefrontal Cortex
Neurons in the prefrontal cortex display sustained activity in response to environmental or internal stimuli, that is continue to fire until the behavioral outcome or a reward signal. Mostly large-scale modeling studies have proposed intensive recurrence and slow excitation mediated by NMDA receptors as crucial mechanisms able to support the sustained excitation in these neurons. In addition, electrophysiological studies suggest that single-cell intrinsic currents also underlie the delayed excitation of prefrontal neurons. This project is focused on the interplay of both the computational and electrophysiological approaches in characterizing the activity observed at layer V prefrontal pyramidal neurons. Towards this goal we use morphologically simplified compartmental models of layer V neurons (both pyramidal and interneurons) implemented in the NEURON simulation environment. These neurons are fully interconnected in a small network, the properties of which are extensively based on anatomical and electrophysiological data.
2. Models of Fear Memory Allocation in the Amygdala
One of the goals of neuroscience is to understand the process via which memories are encoded and stored in the brain. Recent experiments have demonstrated how memories are encoded in specific neuron groups in the brain. Traditionally, it is thought that the strengthening of synaptic connections via synaptic plasticity is the mechanism underlying memory formation in the cortex. New insights indicate that other factors, such as neuronal excitability and competition among neurons may crucially affect the formation of a memory trace. The transcription factor CREB has been shown to modulate the probability of allocation of memory to specific groups of neurons in the Lateral Amygdala. The goal of our computational work is to investigate the process of memory allocation and the properties of the memory trace.
Problems Solved
a) The PFC microcircuit is used to characterize:
- the generation of UP and Down states, observed during both in vivo and in vitro recordings;
- the interplay of single cell ionic with synaptic currents for the emergence of sustained excitability;
- the role of both synaptic and intrinsic plasticity in long term memory formation in the prefrontal cortex.
b) By creating a large scale computational model of the lateral amygdala, we aim to investigate how the modulation of excitability, synaptic plasticity, homeostatic plasticity and neuronal inhibition affect the formation of fear memories in the lateral amygdala.
Scientific and Social Impact
Understanding the properties that make these neurons special in carrying temporal distinct information by using a bottom-up approach is a key issue in unraveling the complicated dynamics and flexibility of prefrontal neurons during behavioral tasks.
Our fear memory simulations can provide insights in the relationships between memory traces and the role of CREB. Our results can be useful in understanding the outcomes of related behavioral and electrophysiological studies.
Collaborations
- Univeristy of Crete - Biology Department
- Buszaki Lab, Rutgers Univ.
Beneficiaries
- Univeristy of Crete - Biology Department
- Computational Biology Lab - IMBB/FORTH
Number of users
2
Development plan
- Concept: Completed.
- Start of alpha stage: M5
- Start of beta stage: M8
- Start of testing stage: M9
- Start of deployment stage: M10
- Start of production stage: M15
Resource requirements
- Number of cores required for a single run: 60
- Minimum RAM/core required: 2GB
- Storage space during a single run: 6GB
- Long-term data storage: 10GB
- Total core hours required: 15000
Technical Features and HP-SEE Implementation
- Primary programming language: NEURON
- Parallel programming paradigm: MPI
- Main parallel code: OpenMPI
- Pre/post processing code: Python and matlab scripts
- Application tools and libraries: NEURON, SciPy, Matplotlib
Usage Example
The application requires compilation of the neuron mechanism libraries in the target platform:
cd APP_DIR/mechanisms; nnrivmodl
The launcher script "start.sh" starts a neuron process with the correct parameters, and it is submitted to the HPC cluster through PBS
mpiexec ../mechanism/x86_64/special -mpi main_code.hoc
Infrastructure Usage
- Home system: HPCG/BG
- Applied for access on: 02.2011
- Access granted on: 02/2011
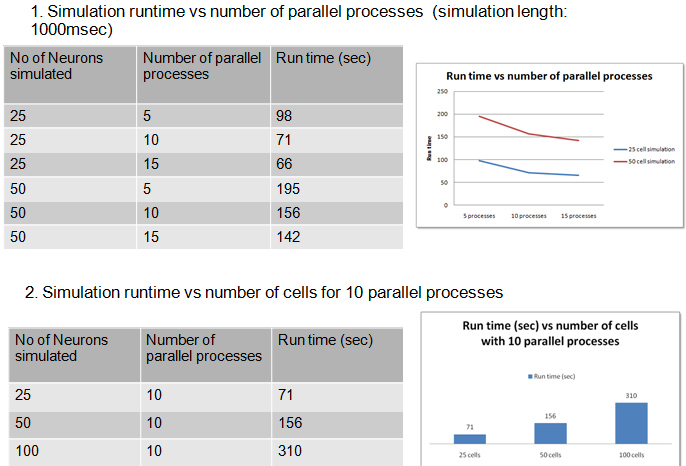
- Achieved scalability: 50 cores
- Accessed production systems:
- Applied for access on: 02.2011
- Access granted on: 02/2011
- Achieved scalability: 50 cores
- Porting activities: Application ported and running since 04/2011
- Scalability studies: 5-20 Cores
Running on Several HP-SEE Centres
Click here for full Benchmark Analysis
- Benchmarking activities and results: Project deployed on HPCG-BG
- Other issues: .
Achieved Results
- Parallel Simulations scaled our existing models up by a factor of 10
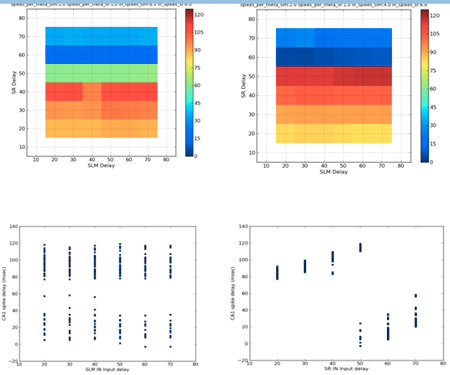
- Extensive parameter exploration of the CA1 stimulation model has been completed
Publications
- Daphne Krioneriti*, Athanasia Papoutsi and Panayiota Poirazi (2011) Mechanisms underlying the emergence of Up and Down states in a model PFC microcircuit. BMC Neuroscience 2011,12 (Suppl 1):O7
- Papoutsi A, Sidiropoulou K and Poirazi P., “Temporal Dynamics underlie bi-stability in a model PFC microcircuit” (submitted)
- Poirazi P. “Dendrites and Information processing” invited seminar, Bernstein Center Freiburg, 27/3/2012
Foreseen activities
- Simulations for larger time frames.
- Simulations with variable connectivity (i.e. not all-to-all networks)
- Assessment of the role of feedback inhibition in sustained activity