MiRs
From HP-SEE Wiki
General Information
- Application's name: Searching for novel miRNA genes and their targets
- Virtual Research Community: Life Sciences
- Scientific contact: Dr. Panayiota Poirazi, poirazi@imbb.forth.gr
- Technical contact: Dr Anastasis Oulas, oulas@imbb.forth.gr
- Developers: Dr Anastasis Oulas, IMBB, FORTH, Greece
- Web site: http://www.imbb.forth.gr/people/poirazi/drupal/?q=node/6/
Short Description
The mode of action of the mature miRNA in mammalian systems is dependent on complementary base pairing to the 3’UTR region of the target mRNA, thereafter causing the inhibition of translation and/or the degradation of the mRNA. Despite the body of evidence supporting a role of miRNAs in numerous molecular processes and diseases, their exact mechanism of action remains to be elucidated because the downstream targets of miRNAs implicated in these prcesses have yet to be defined. Towards this goal, miRNA target prediction tools can offer a first assessment as to which target genes are regulated by miRNAs, thus providing new insights regarding their specific functions and guiding future experiments.
There is great need for a more sophisticated target prediction tool that achieves an inmprved balance between sensitivity and specificity. In order to maximize sensitivity and specificity to the extent that outperforms existing target prediction tools, we implement a new target prediction tool using Hidden Markov Models (HMM). The first step for the successful implementation of this tool is the selection of significant biological features that are implicated in the miRNA::mRNA interaction. Once again careful training and validation has to be performed to obtain statistical evidence for the prediction accuracy of our tool. We have validated our methodology and are now ready to utilize it to scan all human 3’UTR regions for novel miRNA targets. This prediction pipeline will require a great deal of computational power in order to complete these experiments.
Problems Solved
Existing computational miRNA target prediction methods may vary in the algorithm used; and one can state opinions about the strengths or weaknesses of each particular algorithm, the fact of the matter is that all methods fall substantially short of capturing the full detail of physical, temporal, and spatial requirements of biologically significant miRNA::mRNA interactions. The performance of current tools is largely dependent on the overall number of predicted targets (hits). Some tools may be very efficiency in predicting true targets sites (high sensitivity) but at the same time display an extremely large number of overall hits (low specificity). In contrast other tools display an overall high specificity but a very low sensitivity. It is evident that there is great need for a more sophisticated target prediction tool that achieves a balance between sensitivity and specificity. Performing this large scale scan of all miRNAs in the online registry and all the genes of the human genome, will allow us to compare our tool with other available tools on a large scale basis. The results will be stored on an online database that will allow users to query the database using miRNA name or gene name. This will shed some light into new gene targets for miRNAs and the molecular processes that they regulate. Finally experimental verification of computational predictions will be the ultimate step in revealing the molecular pathways of miRNA regulation and characterizing their involvement in disease.
Scientific and Social Impact
The proposed research project is expected to benefit all scientific partners as well as the scientific community of Greece and ultimately contribute towards the enhancement of knowledge in the field of molecular biology of disease. Given the recent establishment of the research field of Computational Biology in Greece, it is anticipated that the proposed project will aid in the creation of a critical mass of scientists with a high level of specialization, capable of conducting quality research work on a multidisciplinary field. We firmly believe that the results expected from the proposed research project will benefit the technological and social development of Greece in many ways. The transfer of knowledge between the research partners will benefit the social development of all sides. The field of miRNAs and cancer is acquiring immense interest in the past few years and constitutes a very hot area of research. The Computaional Biology Lab in IMBB will gain international acknowledgement by conducting research in this field. The publication of research results emerging from this project in internationally renowned journals, will greatly aid towards raising the prestige of IMBB in the scientific community. This will definitely benefit the technological development of Greece and facilitate organization of social networks. This can act as the motivation for Greek researchers working abroad, but also for foreign researchers to consider the possibility of working in Greece in the future. The results arising form the wet lab experiments will enhance the knowledge of the scientific community in the field of miRNAs and cancer. Moreover, it may be possible to select specific targets for therapeutic intervention. In the future such research may act as the stepping stone from which to enhance the health sector of the Greek community.
Collaborations
- Univeristy of Crete - Biology Department
- Univeristy of Crete - Medical Department
- HCMR-IMBG
Beneficiaries
- Univeristy of Crete - Biology Department
- Univeristy of Crete - Medical Department
- HCMR-IMBG
Number of users
1
Development Plan
- Concept: Done before the project started.
- Start of alpha stage: Done before the project started.
- Start of beta stage: M8
- Start of testing stage: M9
- Start of deployment stage: M15
- Start of production stage: M16
Resource Requirements
- Number of cores required for a single run: From 1 to up to 300
- Minimum RAM/core required: 100 MB
- Storage space during a single run: 1 GB
- Long-term data storage: 10 GB
- Total core hours required: 3 000 000
Technical Features and HP-SEE Implementation
- Primary programming language: Java
- Parallel programming paradigm: MPI
- Main parallel code: openMPI
- Pre/post processing code: Java and C
- Application tools and libraries: Targetprofiler, RNAcofold, HMMER
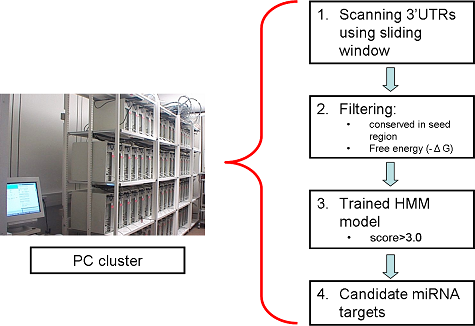
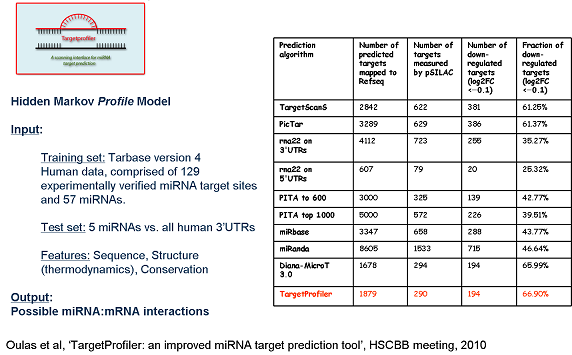
Usage Example
The overall flowchart of the scanning process of Targeprofiler (left panel) as well as a comparison with other available tools (right panel) is shown below.
Infrastructure Usage
- Home system: IFIN_Bio
- Applied for access on: 02.2011
- Access granted on: 02.2011
- Achieved scalability: 256 cores
- Accessed production systems:
- Applied for access on: 02.2011
- Access granted on: 02.2011
- Achieved scalability: 256
- Porting activities: The application has been successfully ported.
- Scalability studies: Tests on 50, 100 and 200 cores.
- Production: Production ready.
Running on Several HP-SEE Centres
- Benchmarking activities and results: At the initial phase the application was benchmarked and optimized on the IFIN_Bio cluster in Romania. After successful deployment on 256 cores the second phase of the benchmarking was initiated and it was deployed on the IFIN_Bio cluster. It was tested there and showed good scalability results on 50, 100 and 200 cores.
- Other issues: .
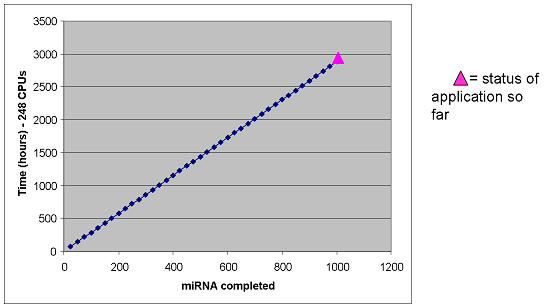
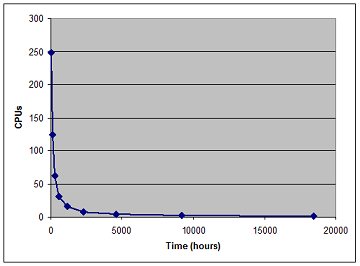
Achieved Results
The final results are shown below (left panel) as well as the CPU usage as a function of real time(right panel). All human miRNAs (~1000) have now been utilized to scan for miRNA targets in all human 3'UTR (~35,000).
Publications
- Oulas, A., Karathanasis, N., Louloupi, A., Iliopoulos, I., Kalantidis, K., Poirazi, P. A new microRNA target prediction tool identifies a novel interaction of a putative miRNA with CCND2 [in preperation].
- Oulas, A., Karathanasis, N., Louloupi, A., Poirazi, P. Finding Cancer-Associated miRNAs: Methods and Tools. Mol Biotechnol. 2011 May 24. [Epub ahead of print]
- Oulas, A., Boutla, A., Gkirtzou, K., Reczko, M., Kalantidis, K., and Poirazi, P. Prediction of novel microRNA genes in cancer-associated genomic regions--a combined computational and experimental approach, Nucleic Acids Res, vol. 37, pp. 3276-87, 2009.
- Oulas, A., M. Reczko, and P. Poirazi, MicroRNAs and Cancer – The Search Begins! Invited review, accepted to IEEE-Transactions on Information Technology in Biomedicine, 2008.
Foreseen Activities
- Message oriented frameworks overcome some deployment limitations like lack of common Grid middleware are installed. This problem is planned to be solved in next step, but it is not an immediate problem for production use.
- The availability of HPC resources enables new research to be performed where we will be investigating the impact of the applied electric field on the devices made from different semiconductor materials.