GENETATOMICS
From HP-SEE Wiki
(→Foreseen Activities) |
|||
| Line 2: | Line 2: | ||
* Application's name: Genetic algorithms in atomic collisions | * Application's name: Genetic algorithms in atomic collisions | ||
| - | * Application's acronym: '' | + | * Application's acronym: ''Genetatomics'' |
* Virtual Research Community: ''Computational Physics'' | * Virtual Research Community: ''Computational Physics'' | ||
* Scientific contact: ''Dragan Jakimovski, dragan.jakimovski@gmail.com'' | * Scientific contact: ''Dragan Jakimovski, dragan.jakimovski@gmail.com'' | ||
| Line 18: | Line 18: | ||
The objective of our work is to calculate these multivariable functions (which arise with Schrodinger equation in general) with different values of parameters and different ranges of variables, which in many cases is computer intensive, and require high amount of CPU time. For now the code has been tested, also on various other systems of equations, prominent in physics, applied mathematics and economics | The objective of our work is to calculate these multivariable functions (which arise with Schrodinger equation in general) with different values of parameters and different ranges of variables, which in many cases is computer intensive, and require high amount of CPU time. For now the code has been tested, also on various other systems of equations, prominent in physics, applied mathematics and economics | ||
* Volter-Lotka system, | * Volter-Lotka system, | ||
| - | * Equations with non-analytical solutions | + | * Equations with non-analytical solutions |
| - | * GDP equations from economic theory | + | * GDP equations from economic theory, and |
| + | * Partial differential equations | ||
For systems for which there is known analytical solution the comparison with the numerically obtained result is excellent. | For systems for which there is known analytical solution the comparison with the numerically obtained result is excellent. | ||
| Line 35: | Line 36: | ||
* Research groups working in atomic / ion collisions and quantum physics in general, for double-checking their own codes; | * Research groups working in atomic / ion collisions and quantum physics in general, for double-checking their own codes; | ||
| - | * Universities and educational | + | * Universities and educational centers with strong program in HP physics computing |
== Number of users == | == Number of users == | ||
| Line 87: | Line 88: | ||
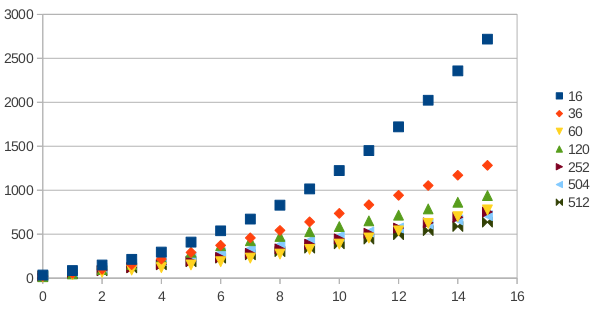
* Benchmarking activities and results: ''The application was successfuly tested for its scalability on HPCG cluster in Bulgaria. Due to long queue waiting times, the problem was not tested for scalability beyond 16 cores. On the HPGCC cluster in Macedonia, the application was used to initially test the system and achieved a scalability of 120 cores. The tests were executed over a medium data set, and we believe that it will scale even more on larger data sets. The results of the scalability measuremets are shown in the following figures:'' | * Benchmarking activities and results: ''The application was successfuly tested for its scalability on HPCG cluster in Bulgaria. Due to long queue waiting times, the problem was not tested for scalability beyond 16 cores. On the HPGCC cluster in Macedonia, the application was used to initially test the system and achieved a scalability of 120 cores. The tests were executed over a medium data set, and we believe that it will scale even more on larger data sets. The results of the scalability measuremets are shown in the following figures:'' | ||
[[File:Genatomics-speedup.png]] | [[File:Genatomics-speedup.png]] | ||
| + | * The inovative approach was adopted for measuring the speed-up of parallel implementation of the algorithm due to its | ||
| + | stochastic nature. With different runs of the code one gets different functions with different evaluation times. To | ||
| + | smooth out this inherent stochasticity of the genetic algorithm we modified the expression for speed-up relative | ||
| + | nonparallel case to | ||
| + | |||
| + | s=(t_j\sum_{i=1}^k <r_{1,i}>) / (t_1\sum_{i=1}^k <r_{j,i}>) | ||
| + | |||
| + | where t_j equals time to develop the n-th generation with j processors, t_1 equals time to develop n-th generation | ||
| + | with one processor, k equals the number of equations in the system, <r_{1,j}> equals the mean value of the stack for | ||
| + | whole population for n-th generation as represents the i-th equation of the system when the algorithm execution | ||
| + | falls on one processor, <r_{j,i}> is the mean value of the stack, for whole population for n-the generation, as | ||
| + | represents the i-th equation of the system when the algorithm runs on j-processors. | ||
| + | With this measure of speeding up, relative serial implementation, the results may be represented on the following | ||
| + | figure: | ||
| + | [[File:Scalability.PNG]] | ||
* Other issues: ''.'' | * Other issues: ''.'' | ||
Revision as of 18:00, 30 September 2012
General Information
- Application's name: Genetic algorithms in atomic collisions
- Application's acronym: Genetatomics
- Virtual Research Community: Computational Physics
- Scientific contact: Dragan Jakimovski, dragan.jakimovski@gmail.com
- Technical contact: "Boro Jakimovski, boro.jakimovski@finki.ukim.mk", Jane Jovanovski, janejovanovski@gmail.com
- Developers: Faculty of Natural Science and Mathematics, UKIM, FYROMacedonia
- Web site: http://wiki.hp-see.eu/index.php/GENETATOMICS
Short Description
The computer simulation and modeling of various processes involving highly charged ions in plasma are extremely important in contemporary physics. The knowledge of cross sections of electron capture and ionization processes as well as of corresponding sections for excitation of electron(s) in atoms and ions colliding with impurity ions in reactor plasma is valuable in calculations of other plasma parameters and diagnostics of its characteristics in experimental reactor ITER to be build in France. Of special interest are the details of cross sections for different plasma temperatures. The usage of HPC for this application will increase the performance of the genetic algorithms, allowing processing of more parameters and larger intervals in single run.
Problems Solved
The objective of our work is to calculate these multivariable functions (which arise with Schrodinger equation in general) with different values of parameters and different ranges of variables, which in many cases is computer intensive, and require high amount of CPU time. For now the code has been tested, also on various other systems of equations, prominent in physics, applied mathematics and economics
- Volter-Lotka system,
- Equations with non-analytical solutions
- GDP equations from economic theory, and
- Partial differential equations
For systems for which there is known analytical solution the comparison with the numerically obtained result is excellent.
Scientific and Social Impact
Contribution to IAEA and atomic collisions research community, as well as research groups in quantum physics in general. The availability of robust, thoroughly tested parallel genetic algorithm code for various quantum problems with different effective potentials should be beneficial to all those developers interested in independent check of their own, more specialised codes. Possible scaling of the code to large number of processors (up to, say 100) for solving more complex problems and eventually to huge number of more than 100 thousand, processors with different hardware / software environment may prove itself to have important social impact.
Collaborations
- International Atomic Energy Agency, Vienna
Beneficiaries
- Research groups working in atomic / ion collisions and quantum physics in general, for double-checking their own codes;
- Universities and educational centers with strong program in HP physics computing
Number of users
3
Development Plan
- Concept: The concept has been planned before the M1.
- Start of alpha stage: M1-M8
- Start of beta stage: M9-M10
- Start of testing stage: M11-M13
- Start of deployment stage: M14-M15
- Start of production stage: M16-M24
Resource requirements
- Number of cores required: 128 - 2048
- Minimum RAM/core required: 0.5GB
- Storage space during a single run: 10MB
- Long-term data storage: 5GB
- Total core hours required: 1 000 000
Technical Features and HP-SEE Implementation
- Primary programming language: FORTRAN
- Parallel programming paradigm: SMP and MPI
- Main parallel code: In-house development
- Pre/post processing code: In-house development
- Application tools and libraries: none, (planned usage of ATLAS, BLAS, GotoBLAS, ScaLAPAC)
Usage Example
The application is currently being used by hardcoding the new problems into the FORTRAN source. The user then recompiles the source and starts it using desired number of processes.
Infrastructure usage
- Accessed production system: HPCG (Bulgaria)
- Applied for access on: 09.2010
- Access granted on: 09.2010
- Achieved scalability: 16 cores
- Porting activities: Moved on Home system
- Scalability studies: Moved on Home system
- Home system: HPGCC.FINKI (Macedonia)
- Applied for access on: 1.2012
- Access granted on: 1.2012
- Achieved scalability: 60 cores for the current data sets (tested on up to 504 cores). The application was tested directly, before the cluster is operational for testing purposes
- Porting activities: ongoing
- Scalability studies: ongoing
Running on several HP-SEE Centers
- Benchmarking activities and results: The application was successfuly tested for its scalability on HPCG cluster in Bulgaria. Due to long queue waiting times, the problem was not tested for scalability beyond 16 cores. On the HPGCC cluster in Macedonia, the application was used to initially test the system and achieved a scalability of 120 cores. The tests were executed over a medium data set, and we believe that it will scale even more on larger data sets. The results of the scalability measuremets are shown in the following figures:
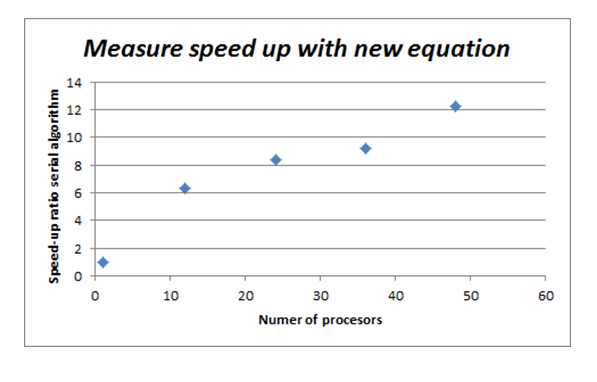
- The inovative approach was adopted for measuring the speed-up of parallel implementation of the algorithm due to its
stochastic nature. With different runs of the code one gets different functions with different evaluation times. To smooth out this inherent stochasticity of the genetic algorithm we modified the expression for speed-up relative nonparallel case to
s=(t_j\sum_{i=1}^k <r_{1,i}>) / (t_1\sum_{i=1}^k <r_{j,i}>)
where t_j equals time to develop the n-th generation with j processors, t_1 equals time to develop n-th generation
with one processor, k equals the number of equations in the system, <r_{1,j}> equals the mean value of the stack for
whole population for n-th generation as represents the i-th equation of the system when the algorithm execution
falls on one processor, <r_{j,i}> is the mean value of the stack, for whole population for n-the generation, as
represents the i-th equation of the system when the algorithm runs on j-processors.
With this measure of speeding up, relative serial implementation, the results may be represented on the following
figure:
- Other issues: .
Achieved Results
Publications
- J Jovanovski, B Jakimovski and D Jakimovski, Parallel Genetic Algorithms for Finding Solution of System of Ordinary Differential Equations, presentation at ICT Innovations 2011 conference
- J Jovanovski, B Jakimovski and D Jakimovski, Improvements of the Parallel Evolutionary Algorithm for finding solution of a system of ordinary differential equations, The 9th Conference for Informatics and Information Technology (CIIT 2012)
- J Jovanovski, B Jakimovski and D Jakimovski, Parallel Genetic Alghorithms for Finding Solution of System of Ordinary Differential Equations, in L. Kocarev (Ed.) ICT Innovations 2011, Advances in Intelligent and Soft Computing, Springer, 2012, Volume 150/2012, 227-237
Foreseen Activities
- Implementation of a hybrid version using OpenMP
- Implementation of a more configurable application that will accept the problem functions using a configuration file without influencing performance
- Making changes in the code to incorporate partial differential equations as well
- Implementation, testing and production of the code for two-heavy-centers-one-electron system